Product Description
These products are ideal for digital PCR and/or Next Generation Sequencing (NGS). In particular:
– Validation and development of sequencing protocols (e.g. Whole Genome Sequencing (WGS), Amplicon Sequencing) and PCR protocols
– Determination of the detection limit of the method
This product contains the following set of tubes:
| Tube | Mutations per tube included |
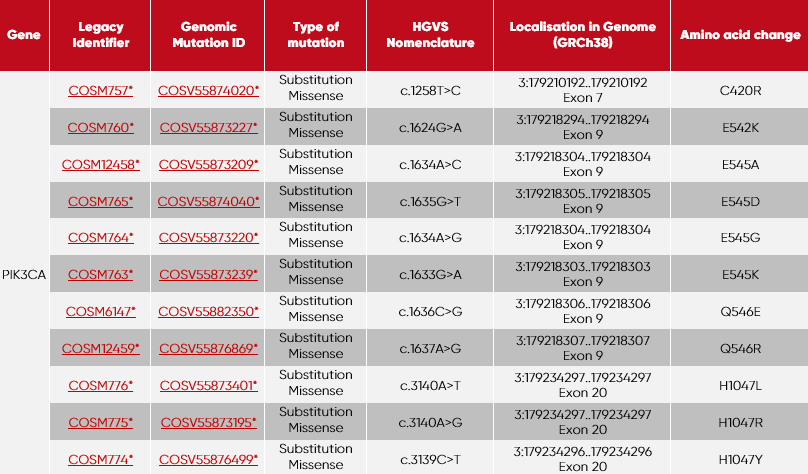
| 1 | PIK3CA-C420R + PIK3CA-E545A |
| 2 | PIK3CA-Q546R + PIK3CA-E542K |
| 3 | PIK3CA-H1047L + PIK3CA-E545D |
| 4 | PIK3CA-H1047R + PIK3CA-E545G |
| 5 | PIK3CA-H1047Y + PIK3CA-E545K |
| 6 | PIK3CA-Q564E |
| 7 | incl. Wildtype (Ashkenazim son cfDNA Cat. No.: SID-000003) |
Each vial has the following features:
Mutations:
Buffer:
Tris-EDTA (10 mM Tris, 1 mM EDTA), pH 8.0
Storage:
2-8 °C
Expiry:
stable for 24 months from date of manufacturing (as supplied)
Quality control
Fragmentation size:
Electrophoresis-Bioanalyzer-High Sensitivity DNA Kit Agilent
Allelic Frequency (metrologically traceable):
ddPCR (Bio-Rad)
Quantification (metrologically traceable):
1. UV-Vis Spectrophotometry (NIST-Reference method)
2. Fluorometric dsDNA measurement (Qubit)
Technical background
Derived from:
The Genome In A Bottle (GIAB) cell line from the Personal Genome Project (PGB): GM24385 (HG002- NA24385 – huAA53E0 – Ashkenazim son)
Bioinformatics:
– Lot specific sequencing files: LOT Search
– High-confidence variant calls: ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/AshkenazimTrio
– Raw datasets and bam files, currently including 10X Genomics, BioNano, Complete Genomics regular and LFR, 300x Illumina paired-end, Illumina 6kb mate-pair, 1000x Ion exome, custom moleculo libraries, ~0.05x Oxford Nanopore, and 70x/30x/30x PacBio: ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG002_NA24385_son/
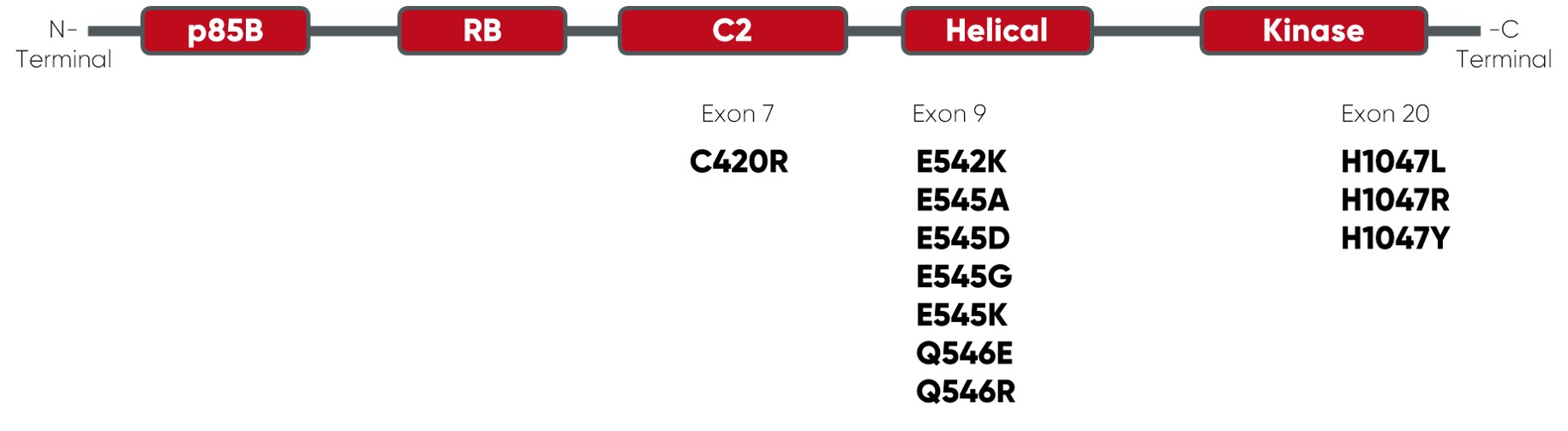
* GRCh38 · COSMIC v90
Literature:
1. The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61-70.
2. Tolaney S, Toi M, Neven P, et al. Presented at: 2019 American Association for Cancer Research (AACR) Annual Meeting; March 29 – April 3, 2019; Atlanta, GA.
3. Di Leo A, Johnston S, Seok Lee K, et al. Lancet Oncol. 2018;19(1):87-100.
4. Moynahan ME, Chen D, He W, et al. Br J Cancer. 2017;116(6):726-730.
5. Mutation distributions and clinical correlations of PIK3CA gene mutations in breast cancer
Indications: For Research Use Only
Manufactured by: SensID GmbH